Leading scientists from academia, industry, and government discuss exciting new developments in diagnostics research
Farid Chehab is professor of Laboratory Medicine at the University of California, San Franscisco (UCSF) and director of the Clinical Molecular Diagnostics Laboratory, UCSF Medical Center. His clinical research interests include the development and implementation of novel molecular diagnostic assays.
Jennifer Leeds is executive director in the Infectious Diseases Area at the Novartis Institutes for BioMedical Research. Her group focuses on the identification of antibacterial targets and discovery of new molecules to address unmet medical needs in patients with severe and drug resistant bacterial infections.
Jean Patel is the Science Team Lead at the Antibiotic Resistance Coordination and Strategy Unit, Centers for Disease Control and Prevention (CDC). Her expertise is in the laboratory detection of antimicrobial resistance.
Ali Yetisen is a Tosteson fellow at Harvard Medical School and Harvard-MIT Health Sciences and Technology. His research involves medical diagnostics including optical sensors, biophotonics, and microfluidics.
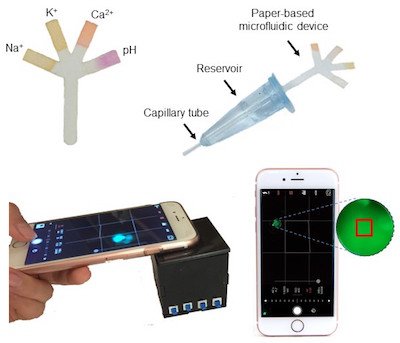
Image credit: Paper-Based Microfluidic System for Tear Electrolyte Analysis. Lab on a Chip 17, 1137-1148 (2017)
Q: What have been the most exciting types of diagnostic strategies developed in the past 5 years?
Leeds: In my field, which is antibiotic discovery and development, I like the idea of the image-based rapid diagnostic system for bacterial infection. It provides a method for quickly determining which bacterial species are present in a patient sample and the susceptibility of those bacterial species to different antibiotics. Currently, it takes 48–72 hours or longer to get a specific diagnosis of a suspected bacterial infection and a clarity about which drugs are appropriate to treat the infection. With the image-based rapid diagnostic system, that information can be gained in just 4–6 hours! And, when a patient is septic, every hour counts.
Yetisen: We have seen significant advances in the development of highly sensitive, quantitative, and multiplex assays. These new technologies include rapid amplicon detection at point-of-care settings and low-cost polymerase chain reaction (PCR) devices and come in the form of disposable paper, plastic, or a combination of the two. We have also seen an increase in the utilization of smartphone devices in the quantification of diagnostic assays, and wearable devices have emerged as a new research front for the continuous monitoring of biomarkers.
Chehab: Undoubtedly, the most exciting and promising DNA diagnostic method for discovery and patient care is currently next-generation sequencing (NGS). This method has already made a significant impact on the diagnosis and management of patients with inherited disorders or cancer, and those afflicted with rare but deadly infectious agents. In addition, digital PCR has been implemented as a quantitative method in clinical laboratories for evaluating the load of mutations such as those occurring in cancer.

Q: Are there other avenues that are being explored that could propel the field of diagnostics into a new era?
Leeds: The handheld device-based diagnostic tools are really interesting. They could help narrow possible pathogens and enable a more rational first choice of therapies, perhaps even at the bedside and in places with limited resources such as in the developing world or at the battlefield. Since these are largely DNA-based technologies at the moment, there will be limitations. Perhaps a breakthrough could come if these technologies were married with a more confirmatory biomarker, such as a metabolite that is specific for certain types of infection.
Yetisen: We are in the middle of a paradigm shift in preventative health monitoring. We are moving toward non-invasive or minimally invasive real-time technologies that can measure analytes in the body over long periods of time. These technologies will create a basis to test individuals before a particular disease progresses and becomes symptomatic.
Chehab: Cell-free DNA testing such as in non-invasive prenatal testing (NIPT) is an area that has really taken off in recent years. The ability to diagnose a fetus without interventions simply from a maternal blood test is currently used for detecting an extra or a missing chromosome (such as for Down syndrome) and is progressing toward single-gene disorders. Similarly, in cancer caused by solid tumors, small amounts of tumor DNA can be found in circulating blood. Analysis of this DNA is referred to as a liquid biopsy, and its purpose is to obviate the need for an invasive tumor biopsy. The typing of the genetic signature of a tumor from a blood sample will have wide ramifications. Imagine, being able to determine the tumor stage of a breast cancer or a glioma without any type of surgery!
Q: Historically, diagnostics and drug treatments have been developing separately. Do you think that it would be beneficial for the two fields to converge?
Disease causing bacteria. Image credit: Antibiotic resistance threats in the United States, 2013 (CDC report)
Patel: When a new antibiotic comes to market, hospital laboratories need a diagnostic to determine if the new drug is active against the bacteria causing a patient’s infection. Too often, the diagnostic test development lags behind, which can result in a new drug being overused or underused. Co-development can also be important when an antibiotic has a very narrow spectrum of activity — for example, the drug is only active for treating infections caused by one species of bacteria. In an era of antibiotic resistance, it could be helpful for patient management if diagnostic tests could quickly identify if a patient is infected with the bacteria or not.
Chehab: It often takes many years between the time a diagnostic test for a genetic mutation is developed and the time a therapy emerges. The DNA-/RNA-based methodologies for diagnostics and therapy are not so disparate, but their complexities are vastly different. For a solid molecular diagnosis, one or a few mutations may be all that is needed. However, in cancer therapy, the emergence of mutations is dynamic and will likely impact prognosis. It is the physician-scientist’s job to eradicate the deleterious DNA variants and maintain the beneficial ones.
Q: We often hear that the holy grail for diagnostics is “anywhere, anytime, anyone.” Is this a reasonable expectation?
Leeds: Reasonable? Yes. Achievable? Hard to say. In some instances, sure. In all instances? Not sure. That should not prevent us from taking a visionary approach to meeting that goal. What we need to pay attention to, though, is the level of training and education that would be necessary to be able to interpret the results. Who is receiving the information? Are they equipped to act on the results? Are the results even actionable? This is what we need to consider as we get closer to this goal.
Patel: We are all hopeful for a portable, real-time test that can help a doctor decide if a patient needs an antibiotic, and tests like this are in development. It will be much harder to develop a bedside test to identify the antibiotic that works best, especially when the infection may be polymicrobial.
Q: Please provide an example of the scientific challenges in developing diagnostics in your field?
Leeds: In the field of novel antibacterial discovery and development, there are several examples of challenges. First, because only a few patient sample-capture methods are truly ‘sterile’ with respect to contamination from normal flora or colonizing non-pathogens, there is a challenge with non-culture-based methods with respect to separating data gathered for the true pathogen versus the non-pathogens. Second, many new diagnostic methods provide information about the potential for drug resistance but don’t provide clear susceptibility data. So — a prescriber may receive a list of what won’t work but still be left wondering what will work! Finally, there are still challenges with matrices (tissue, blood, urine, etc.) and how to develop diagnostics that are agnostic to the matrix, since sample handling needs may differ widely depending on source.
Yetisen: We specifically design and develop diagnostic devices to measure analytes in tear fluid, and there is a void in the basic research of tear biomarker analysis. We have found large range variation and high error margins in the concentration values of tear biomarkers reported in the literature. Such confusion about the biomarker concentration ranges is a problem for device development. Establishing a clear basis for the reference values as well as understanding their possible correlation with blood biomarkers will be helpful in the development of tear analysis diagnostics.
Chehab: NGS generates a lot more information than we can currently handle and interpret. This method has made an impact in analyzing exons, which are regions of the genome that encode proteins and account for only about 1.5% of our genome. So the diagnostic applications of NGS to the whole genome remain daunting because we still don’t understand and cannot interpret variants in many regions of the genome, especially in areas that regulate gene expression. In order to overcome this barrier, we will have to develop rapid and reliable systems to functionally validate suspicious variants that could be pathogenic and therefore diagnostic.
