William G. Kaelin, Jr.
Dana-Farber Cancer Institute, Harvard Medical School

Peter J. Ratcliffe
University of Oxford, Francis Crick Institute

Gregg L. Semenza
Johns Hopkins University School of Medicine
The 2016 Albert Lasker Basic Medical Research Award honors three physician-scientists for their discovery of the pathway by which cells from human and most animals sense and adapt to changes in oxygen availability, a process that is essential for survival. Scientists had long appreciated that the success of today’s dominant life forms hinges on oxygen, yet little was known about their responses to it. William G. Kaelin, Jr. (Dana-Farber Cancer Institute/Harvard Medical School), Peter J. Ratcliffe (University of Oxford/Francis Crick Institute), and Gregg L. Semenza (Johns Hopkins University School of Medicine) illuminated the core molecular events that explain how almost all multicellular animals tune their physiology to cope with varying quantities of the life-sustaining element, thus exposing a unique signaling scheme.
Acceptance remarks
Acceptance remarks, 2016 Lasker Awards Ceremony
I grew up during the space race, so science and engineering were celebrated and supported throughout my childhood and our household had toys that fostered curiosity and creativity, including a microscope and chemistry sets.
In high school, I attended an NSF-sponsored summer program for 32 mathematically gifted students that changed my life. I was delighted to learn that I wasn’t the least gifted of the 32 but I certainly had, due to my abysmal study habits, the lowest grades. I learned that school was more fun when I was challenged, that it is really helpful to be surrounded by people who are smarter than you are, and that I, too, could get good grades if I actually did my homework.
As a premed, I floundered in a research laboratory working on an independent study project that was uninteresting, unimportant, and, as I would later correctly show, undoable. My mentor rewarded me with a “C+,” noting on my transcript that “Mr. Kaelin appears to be a bright young man whose future lies outside of the laboratory.” This painful experience convinced me I was going to be a clinician rather than a scientist, and I approached my clinical training accordingly. I even served as Chief Resident at Johns Hopkins, honing my knowledge of obscure diseases such as von Hippel-Lindau Disease.
Years later, I discovered, thanks to outstanding mentorship from David Livingston, that I actually could do science. And my clinical practice convinced me that we desperately needed better cancer treatments based on a deeper understanding of cancer pathogenesis.
I like mathematics, medicine, and science because I like solving puzzles, and I like answers that are objectively verifiable. Science gathers knowledge, and engineering applies that knowledge to useful purposes. For most of my career, I benefited from the wise decision in our country to have the public sector support basic scientific research, where the timelines and deliverables are too unpredictable for investors, and to let the private sector decide when a line of investigation is ripe for application and commercialization. Although this bargain served American biomedical research well, one hears repeated calls to treat science as though it were engineering, tying funding to anticipated outcomes and impact. This is troubling because many practices that are useful for managing engineering projects are antithetical to good science. For example, building teams and harmonizing goals is often essential for large engineering projects. Early stage science, however, is often driven by creative individuals who follow their curiosity and are willing to go where their science takes them. Forcing scientists into teams and holding them to predetermined deliverables can create herd mentality and stifle the heretical thinking that is often needed for transformative discoveries.
JFK knew it would take a decade to put a man on the moon because it was fundamentally an engineering challenge rather than a scientific challenge. It does scientists and, more importantly, patients and their families, a disservice when fundraisers and policy makers overpromise and oversimplify with respect to our greatest biomedical challenges, including cancer.
I am very proud to receive this award on behalf of the talented young scientists who have worked in my laboratory over the years, to share it with such esteemed colleagues, and to dedicate it to my beloved wife Carolyn, who died last summer.
Acceptance remarks, 2016 Lasker Awards Ceremony
I am deeply honoured to receive the Lasker Foundation Award for Basic Medical Research today.
I’d like to reflect for a moment on the many twists and turns that brought me to this fortunate position. One still clear in my mind dates to the Lancaster Royal Grammar School, circa 1970. I was a tolerable schoolboy chemist and intent on a career in industrial chemistry. The ethereal (but formidable) Headmaster appeared one morning in the chemistry classroom. ‘Peter’ he said with unnerving serenity ‘I think you should study medicine’. And without further thought, my university application forms were changed. To this day, I am unsure whether he felt I would be a good doctor or a bad chemist. But the experience is (I think) a reminder of the role of serendipity in a scientific career, at least in mine.
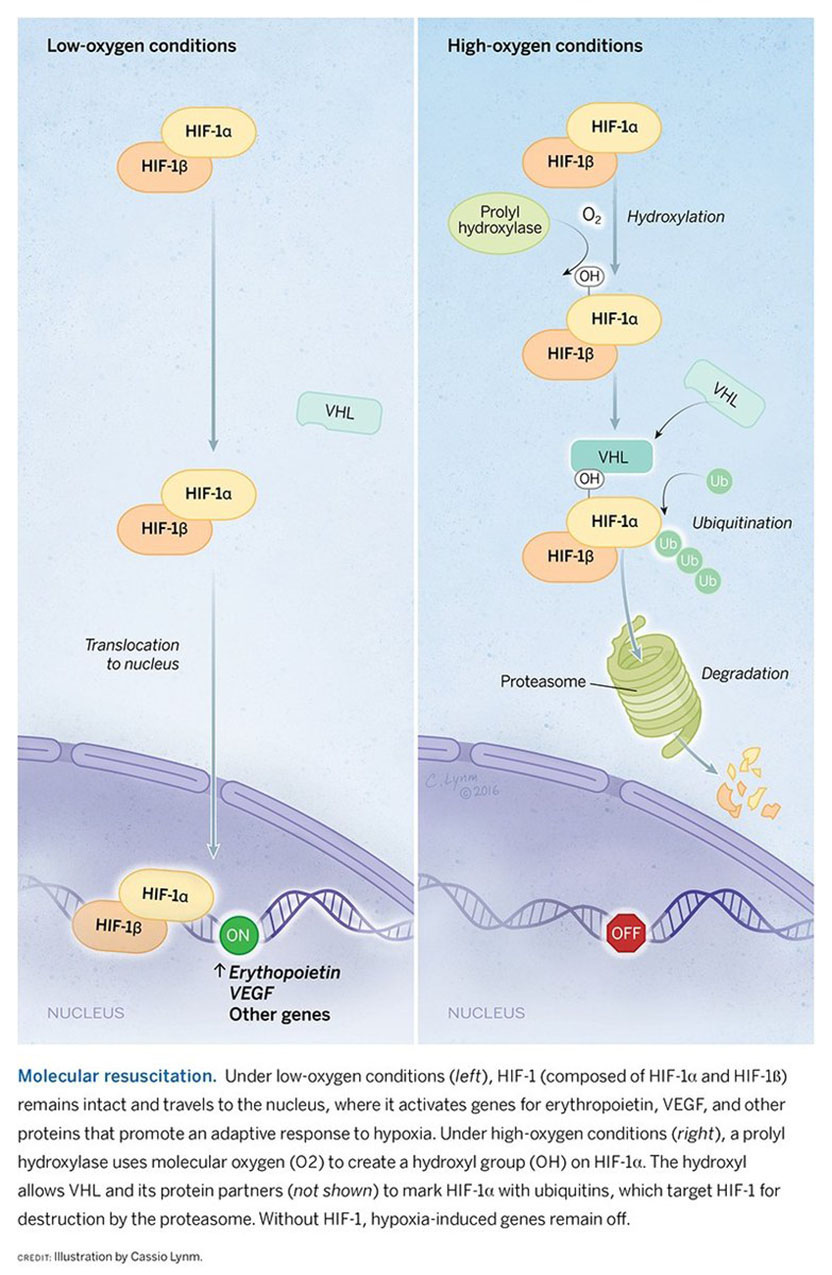
I did train in medicine, as a nephrologist, and came to research rather late, fascinated by the extraordinary sensitivity with which the kidneys regulate the hormone erythropoietin in accordance with blood oxygen content. I felt that the process was interesting and might be tractable. At the time, the erythropoietin gene had recently been identified, so there was a new opportunity for study. But some felt, with the emerging success of recombinant erythropoietin treatment, that understanding how the hormone was regulated was niche area, unlikely to be of general importance.
Acceptance remarks, 2016 Lasker Awards Ceremony
Many of us who conduct biomedical research do so with what could be described as a religious fervor. This would not have come as a surprise to Mary Lasker. She once told a reporter, “I am opposed to heart attacks and cancer the way one is opposed to sin.”
Amen.
Seven months ago, Antonin Scalia died. He had a heart attack, which occurs when the flow of blood through one of the coronary arteries is blocked, cutting off the heart muscle’s supply of oxygen.
Eight months ago, David Bowie died of liver cancer. Cancer cells invade surrounding tissue, make their way into blood vessels, and spread throughout the body. What are they searching for? My guess is oxygen.
Over the last 25 years, we’ve worked to understand how a family of proteins, which we called hypoxia-inducible factors, controls the ability of cells, tissues, and organ systems to respond to changes in oxygen availability. Our work began with an attempt to figure out how expression of the erythropoietin gene was turned on when the body was deprived of oxygen. Now we know that more than 2,000 other genes are regulated in a similar manner.
Looking forward, I’d like to think that within the next decade drugs that stimulate hypoxia-inducible factors will be used to treat anemia and cardiovascular disease, while drugs that inhibit these factors will prolong the lives of cancer patients.
So this is my religion: I am filled with Wonder at the outcome of 4 billion years of evolution here on our speck in the universe and Hope regarding our opportunity to improve the lives of those around us through basic science discoveries and their translation to clinical practice.
Well, I began with Mary Lasker, so I’ll end with Albert. As you know, Mr. Lasker was a successful advertising executive. He was in charge of accounts for products like Sunkist oranges and Pepsodent toothpaste before he and Mary began their crusade to promote biomedical research. Coincidentally, my grandfather also worked for an advertising agency here in Manhattan. His account was for a product called Wonder Bread.
Thank you.
The 2016 Basic Award video
Video Credit: Flora Lichtman