C. David Allis
The Rockefeller University

Michael Grunstein
University of California, Los Angeles
For discoveries elucidating how gene expression is influenced by chemical modification of histones—the proteins that package DNA within chromosomes.
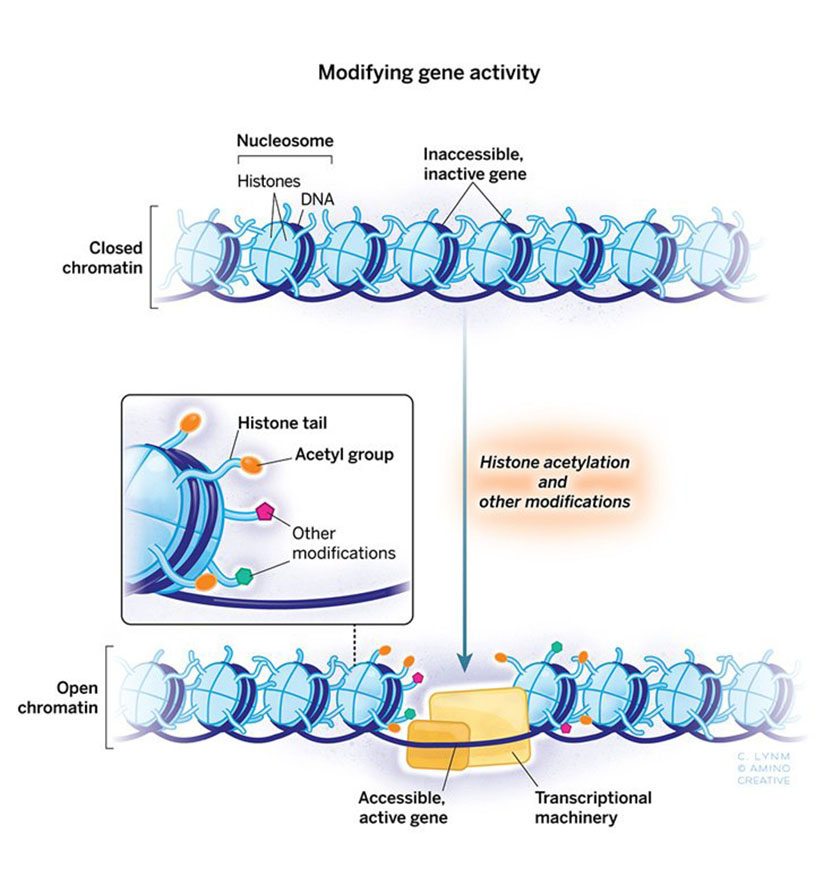
The 2018 Albert Lasker Basic Medical Research Award honors two scientists for discoveries that have elucidated how gene expression is influenced by chemical modification of histones, the proteins that package DNA within chromosomes. Through tour-de-force genetic studies in yeast, Michael Grunstein (University of California, Los Angeles) demonstrated that histones dramatically influence gene activity within living cells and laid the groundwork for understanding the pivotal role of particular amino acids in this process. C. David Allis (Rockefeller University) uncovered an enzyme that attaches a specific chemical group to a particular amino acid in histones, and this histone-modifying enzyme turned out to be an established gene co-activator whose biochemical capabilities had eluded researchers. Grunstein and Allis unveiled a previously hidden layer of gene control and broke open a new field.

What Makes a Piece of Art or Science a Masterpiece?
Critics of art and philosophers of science have long wrestled with the question of what elevates a piece of art or a set of experiments to masterpiece status.
Award presentation by Richard Lifton
The very existence of life seems miraculous. The development from a single cell of a plant, an insect, or a human that each contains specialized cells and organs with radically different functions represents one of the great mysteries. We now understand that all life forms on earth use information encoded in DNA as the instruction manual for building an adult organism. Instructions are executed by copying segments of DNA, called genes, into RNA copies, that direct the production of the proteins that create a muscle cell, liver cell or neuron. This requires that different sets of genes are turned on, turned off, or tuned to the right levels in different cell types. Moreover, gene expression must also must be modulated to respond to periodic changes in the external environment.
Our first insights into how genes are selectively turned on and off came from studies of the bacterium E. coli by Jacob and Monod in the early 1960s. They established the paradigm that transcription factors- proteins that bind to specific DNA sequences- promote or inhibit the copying of DNA into RNA.
Acceptance remarks
Acceptance remarks, 2018 Lasker Awards Ceremony
What could be more “basic” than: 1) biomedical research—the belief that a mechanistic understanding of fundamental problems in biology will lead to translational advances, and 2) histone proteins—the proteins that serve to package our genome. I am a true believer in both. As an undergraduate I was exposed to doing “real experiments”, which, ironically, involved chasing elusive molecules in a cold room. This was a total turn on to me, and I quickly abandoned all thoughts about medical school. I wanted to tackle something at the heart of most biological processes—something in the nucleus of the cell—ideally working on gene regulation. I became intrigued with the biology of a pond water critter called Tetrahymena. A single-celled organism, Tetrahymena runs its life with two genomes partitioned in separate nuclei. One genome is kept in an active state, keeping the cell alive and well—perfect for exploring histone proteins. I was hooked.
The general view that histone modification was a critically important ‘switch’ for gene activation was championed more than 50 years ago by Vincent Allfrey who did all of his work at The Rockefeller University. But who put these chemical flags on; who took them off? This became the long-term goal of my lab, and Tetrahymena was going to be my secret weapon. I had no plan B, and nothing could have been farther from my mind than a Lasker Award.
Not everybody is cut out for science. Fortunately for me, Jim Brownell was. Jim was a graduate student in my lab some 25 years ago, and he is with us here today. Jim worked his magic in the cold room finding the gene switch that Allfrey had suggested might exist. Histones were doing more than just packaging of our genes—tthey were active participants in gene regulation, and Jim had found a protein switch that acted to turn them on. The implications of these findings were immediate and far-reaching. Histone modification was important; Allfrey had it right. Moreover, many of these proteins have proven to be druggable and some are FDA-approved with more on the horizon. These drugs are helping cancer patients lead healthier lives. Basic research and basic histones do matter.
To conclude, I never expected to win a Lasker Award, but I did always know that I had an amazing supporting cast. And as much as I like “histones”, I feel in love with a “herstone.” I thank my wife and family for all of their support. In 1997, Jim himself won a prize for his thesis and was interviewed after his prize ceremony. He commented, “Science is so funny. You are really famous among about a dozen people.” Jim said it right. We do what we do because it is interesting. It is a wonderful journey that I am privileged to take every day.
Congratulations to all of this year’s Lasker Awardees. Thank you all for this remarkable honor.
Acceptance remarks, 2018 Lasker Awards Ceremony
It is very humbling and a great honor to have been chosen for this award by the Lasker Jury and Foundation. I’m particularly pleased to share the honor with David Allis, whose contributions to the field of histone function have been pioneering.
And while David and I have the privilege of accepting the award, we do so in recognition that the Lasker not only awards the individual, but also the field of histone function. This area did not develop in a vacuum. We accept this award knowing that we have benefited from enormous advances in technology that began in the 1970s and that continue today. Our understanding of biology has always been linked to the progress of technology and that symbiosis has become so fruitful that the inventors could never have imagined the sophisticated extensions of their original findings
That is the history of this scientific discipline. It emphasizes that basic science of model microorganisms, funded mainly by the NIH, has led to the establishment of one of the most active fields in the biological and medical sciences and in the world-wide industry of biotechnology. Still, I believe that our understanding of the relevance of histone function is in its infancy. Histone associated proteins may prove themselves to be targets for novel pharmaceuticals. And technologies like gene editing may be able to reverse the defective histone modifications that are associated with certain disease states. David Allis will mention one such histone modification associated with pediatric brain cancer. There is even hope that reversals will one day be possible for certain neurological disorders, like RETT syndrome, given the finding that even advanced RETT symptoms are reversible in mouse models.
I consider myself extremely fortunate to have entered the field when I did. Was there a “eureka” moment? There was. It was when we replaced histone genes with certain mutant versions and saw these mutant histones incorporate into the chromatin of living yeast cells. Once we had a model system, our lab and others in the field could make changes, even in the most conserved regions of histone proteins, and ask new questions regarding novel histone functions. For instance, how can the same histones be involved both in gene activation and in gene repression?
I’ve struggled with the question of how to advise young scientists, since there is no one path to scientific discovery and it is often the discovery of the path itself that makes us so excited to be in Science. But at the expense of stating the obvious, the scientific questions we try to answer should be sufficiently bold so that we can invest ourselves with a focus, persistence and passion to address them.
Again, my thanks to the Lasker Foundation for this honor and to my family, friends and colleagues who have joined us here today.