Alec Jeffreys
University of Leicester

Edwin M. Southern
University of Oxford
For development of two powerful technologies— Southern hybridization and DNA finger-printing — that together revolutionized human genetics and forensic diagnostics.
The 2005 Albert Lasker Award for Clinical Medical Research honors two scientists who revolutionized human genetics and forensic diagnostics. By inventing a method for detecting specific DNA sequences amidst the huge genomes of complex organisms, Edwin Southern infused genetic analysis with tremendous power. Suddenly scientists could study genetic variation in detail and decipher gene structures. Using this technology, Alec Jeffreys devised 'genetic fingerprinting', a way to distinguish every person from every other person, except an identical twin. Its ability to establish family relationships as well as individual identity has helped solve crimes, settle paternity and immigration disputes, establish the bases of inherited diseases, enhance transplantation biology, save endangered species, establish human origins and migrations, and advance countless other beneficial endeavors.
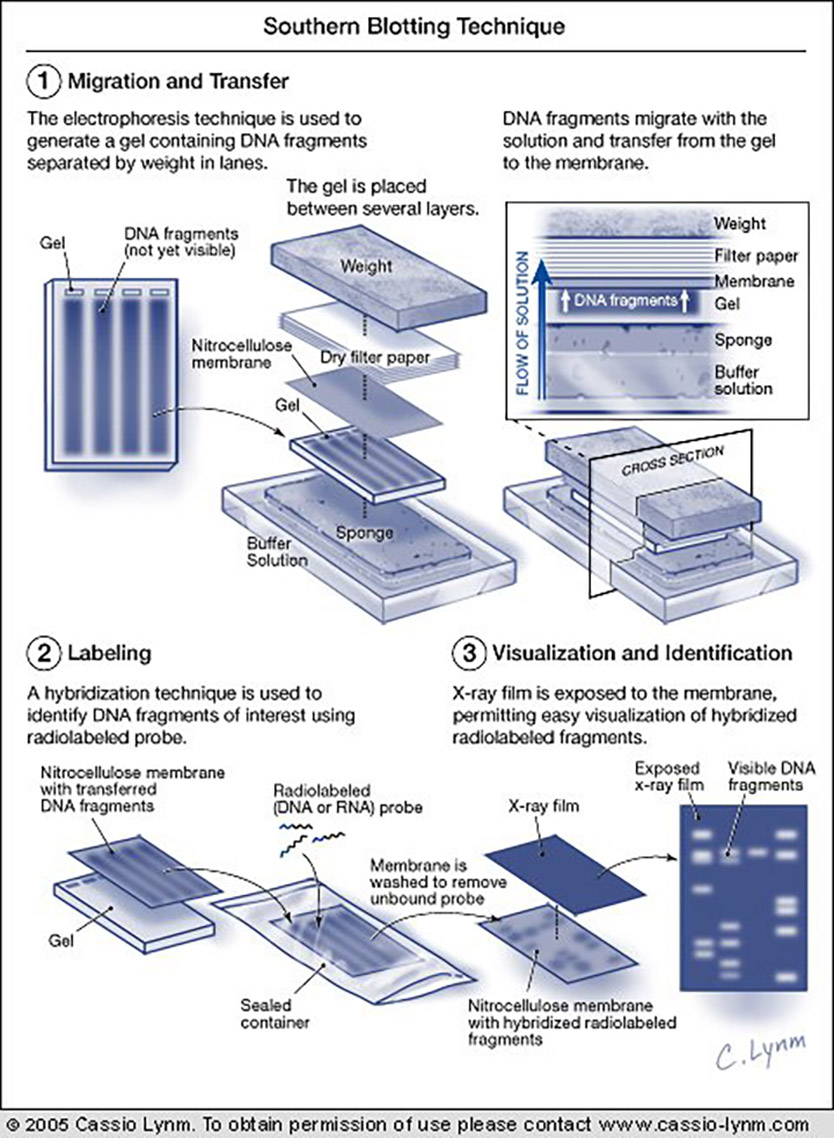
Technology has always defined the strength of genetic analysis. Until the mid-1970s, the ability to locate most genes or sequences of interest on the chromosomes of complex organisms was nearly impossible. This situation severely restricted efforts to define genetic differences that characterize species, individuals, and specific cell types, thus hampering the study of subjects as diverse as evolution and the physiological characteristics of distinct tissues.
Award presentation by Joseph Goldstein
 In the last 50 years, the world has been radically changed by three inventions — burgers, chips, and genes. Let me explain. First, the burger. I ate my first Big Mac in 1966 while I was a resident at the Massachusetts General Hospital in Boston. I vividly remember telling Michael Brown, a fellow house officer at the time, about my fabulous epicurean experience, and for the past 37 years Mike and I have had many exciting discussions about cholesterol while devouring Big Macs.
In the last 50 years, the world has been radically changed by three inventions — burgers, chips, and genes. Let me explain. First, the burger. I ate my first Big Mac in 1966 while I was a resident at the Massachusetts General Hospital in Boston. I vividly remember telling Michael Brown, a fellow house officer at the time, about my fabulous epicurean experience, and for the past 37 years Mike and I have had many exciting discussions about cholesterol while devouring Big Macs.
Since the early 1960s, McDonalds has grown from a few restaurants in California to 30,000 restaurants in 120 countries.
Every day, 10 percent of Americans eat at McDonalds. McDonalds owns more real estate than any other entity in the world — the Catholic church included. Ray Kroc founded McDonalds. His ingenious idea was that people wanted to be served in 60 seconds. McDonalds is a cogent illustration of how one good idea by one person can change the way we live — for better or worse! Or, to paraphrase Woody Allen, "This is the transmutation of life — by a lowly hamburger."
Acceptance remarks

Acceptance remarks, 2005 Lasker Awards Ceremony
The Foundation has done me a great, and I have to say wholly unexpected, honor today, and I am delighted to accept this award not only for myself but also on behalf of all my colleagues who helped turn my dream of DNA identification 20 years ago into a technology that has reached out and directly touched the lives of millions of people worldwide.
Our discovery of DNA fingerprinting was of course totally accidental — what we were really trying to do at the time was to find highly informative DNA markers for basic genetic analysis in humans.
But at least we had the sense to realize what we had stumbled upon. At the time, 20 years ago, we saw the potential for DNA maybe in the occasional specialist forensic or civil law case. What has staggered me is just how dramatic the spread of DNA testing has become. Let me just give two examples. First, the United Kingdom National DNA database now contains the DNA profiles of 3 million UK citizens, which on a proportionate basis would mean that, were we in the United Kingdom, then a dozen or so people in this room would be on that database. Second, I recently had lunch with senior judges at the Old Bailey courts in London and met one judge who was really excited as he was trying a case which, for a change, did not involve any DNA evidence. And it’s not just crime — for example, DNA is clearly going to play a major role in identifying the victims of the Hurricane Katrina disaster.
The impact of DNA fingerprinting has been extraordinary in terms of sheer numbers of people tested. But these bald numbers hide countless personal stories — some squalid, some touching, some tragic. Perhaps one of the most inspiring is that of Kirk Bloodsworth, a truly courageous man whom I am proud to count as a personal friend. He was convicted of a terrible murder back in 1984 and served nine years in a US penitentiary, including two years on death row, before DNA testing exonerated him and led to the identification of the true murderer. Kirk is now a tireless campaigner for post-conviction DNA testing, one of the most important aspects of DNA fingerprinting.
I am very proud of DNA fingerprinting and of the effect that it has had on society and criminal justice. My baby has come of age and I am pleased to say it’s flourishing. I thank you all for honoring it.

Acceptance remarks, 2005 Lasker Awards Ceremony
My big contribution to science was the discovery that blotting paper could be used to soak liquid out of jelly. I wouldn’t be here today if others had not found clever applications for this simple discovery and I’m delighted to share this prestigious award with Alec Jeffreys who used the method to make the most important discovery that genes have a split structure — the introns in eukaryotic genes — and he also developed his famous fingerprinting method from it. But there have been others. I get a little bit of credit for each application, as there have been many, colleagues have been generous in their acknowledgement, and the credit has mounted up over the years. I hope that anyone who has used the method, learning of this award, will feel that they have earned a share in it.
The molecular sciences have brought wonderful progress to biology and medicine in recent decades. It has been a privilege to have lived through this period and to have played a part in it. But this is a beginning. There is much to be done now that we have easy means to read gene sequences and to exploit them. Progress will need new methods, but methods development is not always given the recognition it deserves. I hope that this prestigious award from the Lasker Foundation will encourage others to engage in this most rewarding aspect of science.